7 Niche annotation using KNN
Load the psuedobulked cell type references
# A function to get the niche counts out of the Seurat objects
grab_nichecounts <- function(xenium_object, k_param, cell_types, fov ="fov"){
# Steal from the Seurat BuildNicheAssay function to get nearest neighbours
coords <- GetTissueCoordinates(xenium_object[[fov]], which = "centroids")
cells <- coords$cell
rownames(coords) <- cells
coords <- as.matrix(coords[, c("x", "y")])
neighbors <- FindNeighbors(coords, k.param = k_param)
ct.mtx <- matrix(data = 0, nrow = length(cells), ncol = length(unlist(unique(xenium_object[[cell_types]]))))
rownames(ct.mtx) <- cells
colnames(ct.mtx) <- unique(unlist(xenium_object[[cell_types]]))
cts <- xenium_object[[cell_types]]
for (i in 1:length(cells)) {
ct <- as.character(cts[cells[[i]], ])
ct.mtx[cells[[i]], ct] <- 1
}
niche_counts <- as.matrix(neighbors$nn %*% ct.mtx)%>%t()
niche_counts[1:5,1:5]
return(niche_counts)
}Get each cell’s 50 neareast neighbours
p1_nichecounts_detailed <- grab_nichecounts(cropped.obj, 50,cell_types = "MMR_atlas_lowlevel_pred", fov = "zoom")
p1_nichecounts_detailed[1:5,1:5]
#> bggahoon-1 bggaimpb-1 bggalkpc-1
#> cE01 (Stem/TA-like) 17 18 17
#> cE03 (Stem/TA-like prolif) 30 31 29
#> cE11 (Enteroendocrine) 0 0 0
#> cM06 (DC IL22RA2) 0 0 0
#> cTNI24 (NK GZMK+) 0 0 0
#> bggbeool-1 bggbnono-1
#> cE01 (Stem/TA-like) 18 16
#> cE03 (Stem/TA-like prolif) 30 28
#> cE11 (Enteroendocrine) 0 0
#> cM06 (DC IL22RA2) 0 0
#> cTNI24 (NK GZMK+) 0 0Variable features (cell types) in the data
combined_niche_seu <- CreateSeuratObject(p1_nichecounts_detailed)
combined_niche_seu <- NormalizeData(combined_niche_seu, scale.factor =1)
combined_niche_seu <- FindVariableFeatures(combined_niche_seu)
top20 <- head(VariableFeatures(combined_niche_seu), 20)
plot1 <- VariableFeaturePlot(combined_niche_seu)
plot2 <- LabelPoints(plot = plot1, points = top20, repel = TRUE)
plot2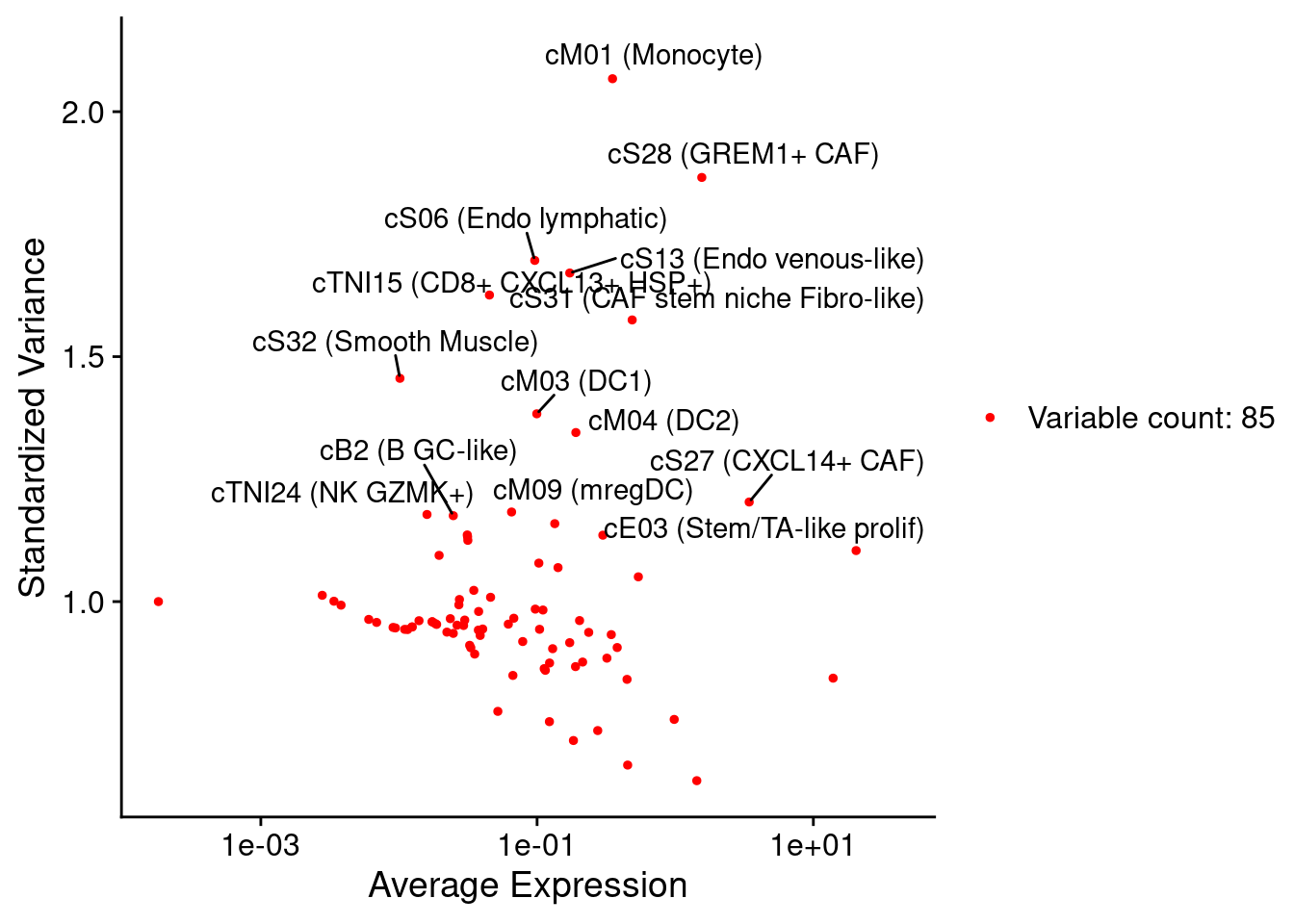
Elbow plot of niche-level PCA
combined_niche_seu <- ScaleData(combined_niche_seu)
combined_niche_seu <- RunPCA(combined_niche_seu, features = VariableFeatures(object = combined_niche_seu))
DimPlot(combined_niche_seu, reduction = "pca") + NoLegend()
ElbowPlot(combined_niche_seu,ndims = 30)+
geom_vline(xintercept = 5, colour="blue", linetype = "longdash")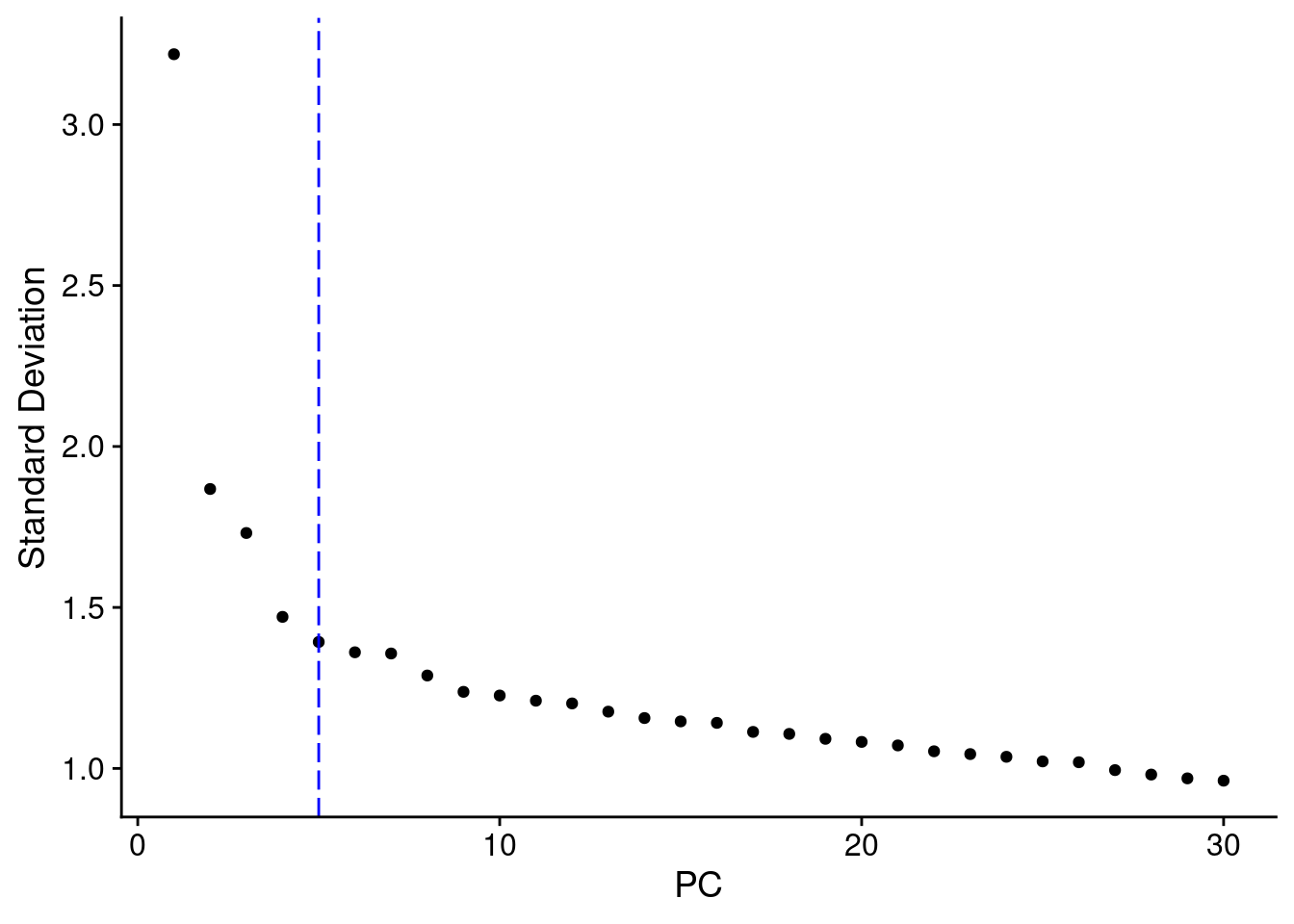
UMAP of KNN matrix
# Even running this with 10 PCs look ok, but maybe a bit complicated
combined_niche_seu <- FindNeighbors(combined_niche_seu, dims = 1:5)
# Setting a low resolution so we don't end up with an unmanageable number of niches
combined_niche_seu <- FindClusters(combined_niche_seu)
#> Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
#>
#> Number of nodes: 16537
#> Number of edges: 449088
#>
#> Running Louvain algorithm...
#> Maximum modularity in 10 random starts: 0.9618
#> Number of communities: 64
#> Elapsed time: 1 seconds
combined_niche_seu <- FindClusters(combined_niche_seu, resolution = 0.1)
#> Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
#>
#> Number of nodes: 16537
#> Number of edges: 449088
#>
#> Running Louvain algorithm...
#> Maximum modularity in 10 random starts: 0.9869
#> Number of communities: 36
#> Elapsed time: 1 seconds
combined_niche_seu <- RunUMAP(combined_niche_seu, dims = 1:5)
DimPlot(combined_niche_seu, reduction = "umap", group.by = "seurat_clusters", label = T)+NoLegend()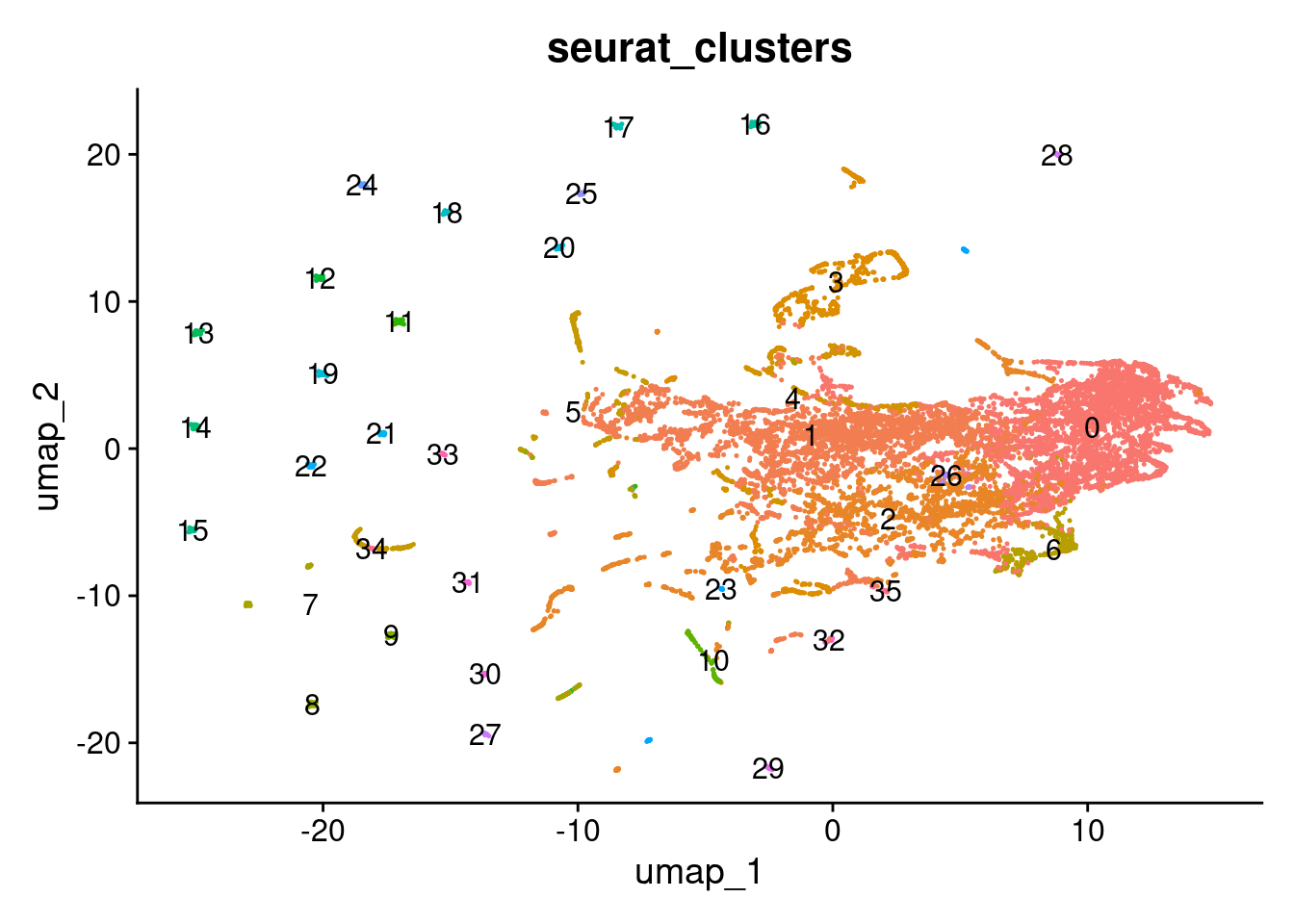
marks <- FindAllMarkers(combined_niche_seu)Plot the niches on the slide
# Check everything lines up
all.equal(colnames(combined_niche_seu), colnames(cropped.obj))
#> [1] TRUE
cropped.obj$Niche <- combined_niche_seu$seurat_clusters
# Plot the 42 predicted niches
plot <- ImageDimPlot(cropped.obj,fov = "zoom", group.by = "Niche",
size = 0.3, cols = "polychrome",
dark.background = T, boundaries = "segmentation",border.size = 0.1,
nmols = 20000) +
ggtitle("Niches")
plot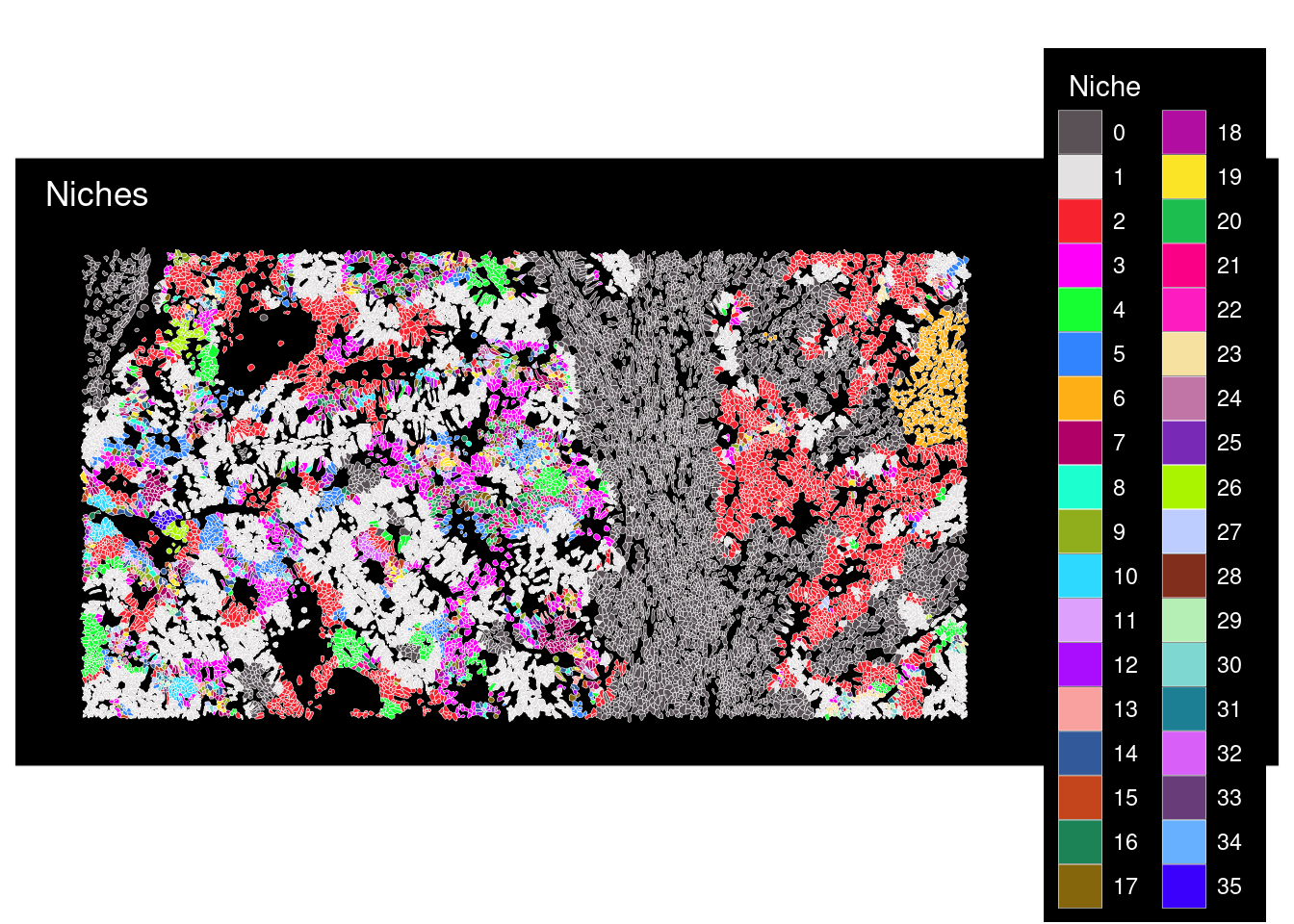
7.1 Plot the niche composition in terms of cell type makeup
niche_makeup_gut <- cropped.obj@meta.data %>%
group_by(Niche) %>%
mutate(Total = n()) %>%
group_by(MMR_atlas_midlevel_pred, Niche, Total) %>%
summarise(Count = n(), .groups = "drop") %>%
mutate(Niche_pct = Count / Total * 100) %>%
arrange(Niche) %>%
mutate(Niche = factor(Niche, levels = unique(Niche)))
# Auto name the niches
niche_names <- niche_makeup_gut %>%
group_by(Niche) %>%
arrange(desc(Niche_pct), .by_group = TRUE) %>%
slice_head(n = 3) %>%
mutate(label = paste0(MMR_atlas_midlevel_pred, " (", round(Niche_pct), "%)")) %>%
summarise(Niche_name = paste(label, collapse = " – "), .groups = "drop")
niche_makeup_gut <- niche_makeup_gut %>%
left_join(niche_names, by = "Niche")
# Create a mapping vector from cluster to name
niche_name_vec <- setNames(niche_names$Niche_name, niche_names$Niche)
# Assign to metadata
cropped.obj$Niche_named <-as.character(niche_name_vec[as.character(cropped.obj$Niche)])
ggplot(data = niche_makeup_gut, aes(x = Niche, y = Niche_pct, fill = MMR_atlas_midlevel_pred))+
geom_bar(stat = "identity")+
labs(x = "Niche", y = "Celltype % of niche", fill = "Cell type")+
blank_theme+
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))