12 Introduction
For today’s session, we will be using the R statistical programming language to perform functional enrichmemt analysis using a range of dedicated R packages. To simplify our work, we will be using the RStudio integrated development environment.
RStudio provides a central place to write code, comments, run code, and view output. We will be using the workshop VMs today which run RStudio and have all the required R packages pre-installed.

12.1 Day 2 overview
- R environment setup including VM login
- ORA with
gprofiler2including multi-query of up and down regulated genes - GSEA over KEGG database with
clusterProfilerand multiple enrichment visualisations withenrichplot - ORA and GSEA with
WebGestaltR, interactive HTML reports, and exploration of term redundancy options - Novel species FEA with
clusterProfiler,WebGestaltRandSTRING(web) - End of workshop summary
12.2 R FEA packages for FEA
There are numerous R packages for FEA, each with their strenghts and limitations. It was difficult to restrict the chosen tools in order to fit within the time allocated for the workshop!
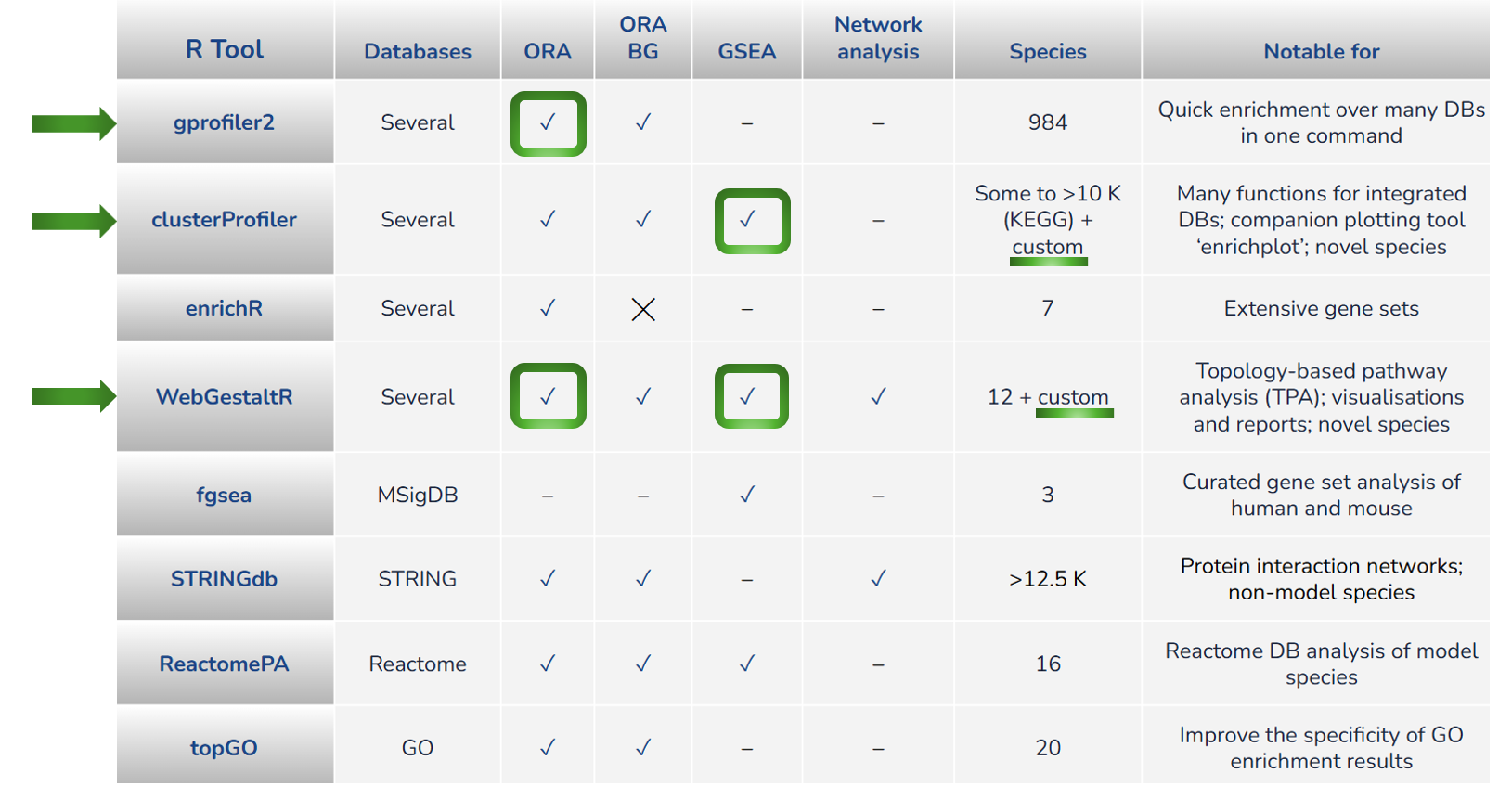
The following three R tools have been selected for today’s workshop:
-
gprofiler2due to its ease of use, high number of supported species, and multiple database enrichments produced within a single run. Caveat: only ORA analysis -
clusterProfiler2due to its integrated database support, runs both GSEA and ORA, companion plotting toolenrichplotwith diverse plot options, and novel species support -
WebGestaltRdue to its ease of use, high number of suported databases and namespaces, runs both GSEA and ORA, interactive HTML reports and plots, and novel species support
12.3 Day 2 approach
For this part of the workshop, we will be doing a semi code-along approach, where you are not required to type any code (apart from a few basics in the RStudio familiarisation part next up) but we will be running code chunks together, talking about what each part of the code is doing, and viewing the output in real time.
All of the code required has been prepared into separate code files for each activity, and we will download these to the VMs in the next section.