10 Resources
The following tools might be useful for downstream functional analysis; this includes some not covered in todays workshop.
Note that most of these tools do more than just enrichment tests, and some include their own databases.
10.1 g:Profiler / g:GOSt
https://biit.cs.ut.ee/gprofiler/gost
The g:GOSt ‘functional profiling’ tool of gProfilercalculates functional enrichment for gene lists, identifying enriched terms across various ontologies and pathways.
It features a clean, modern interface and provides a handy summary highlighting which genes contribute to the enrichment.
Figure 10.1: The gProfiler front page
10.2 PANTHER
PANTHER performs overrepresentation tests across multiple databases; Gene ontology, reactome, PANTHER pathways and protein classes. Allows more control over the statistical test used and clearly summarises what was actually done.
Figure 10.2: PANTHER
10.3 DAVID
Via its ‘functional annotation’ tool, DAVID allows you to calculate functional enrichment across a number of databases ; Gene Ontology, KEGG, reactome and others. Reliable, with a slightly clunky interface.
Figure 10.3: DAVID
10.4 Enrichr
https://amp.pharm.mssm.edu/Enrichr/
Enrichr easily calculates enrichment across a wide range of databases. Currently does allow for a background set.
Figure 10.4: Enrichr
10.5 Reactome
The core of reactome is the reactome pathways and browser. ALthough other tools use the reactome database, the reactome website provides a means to browse enrichment within the pathway browser view.
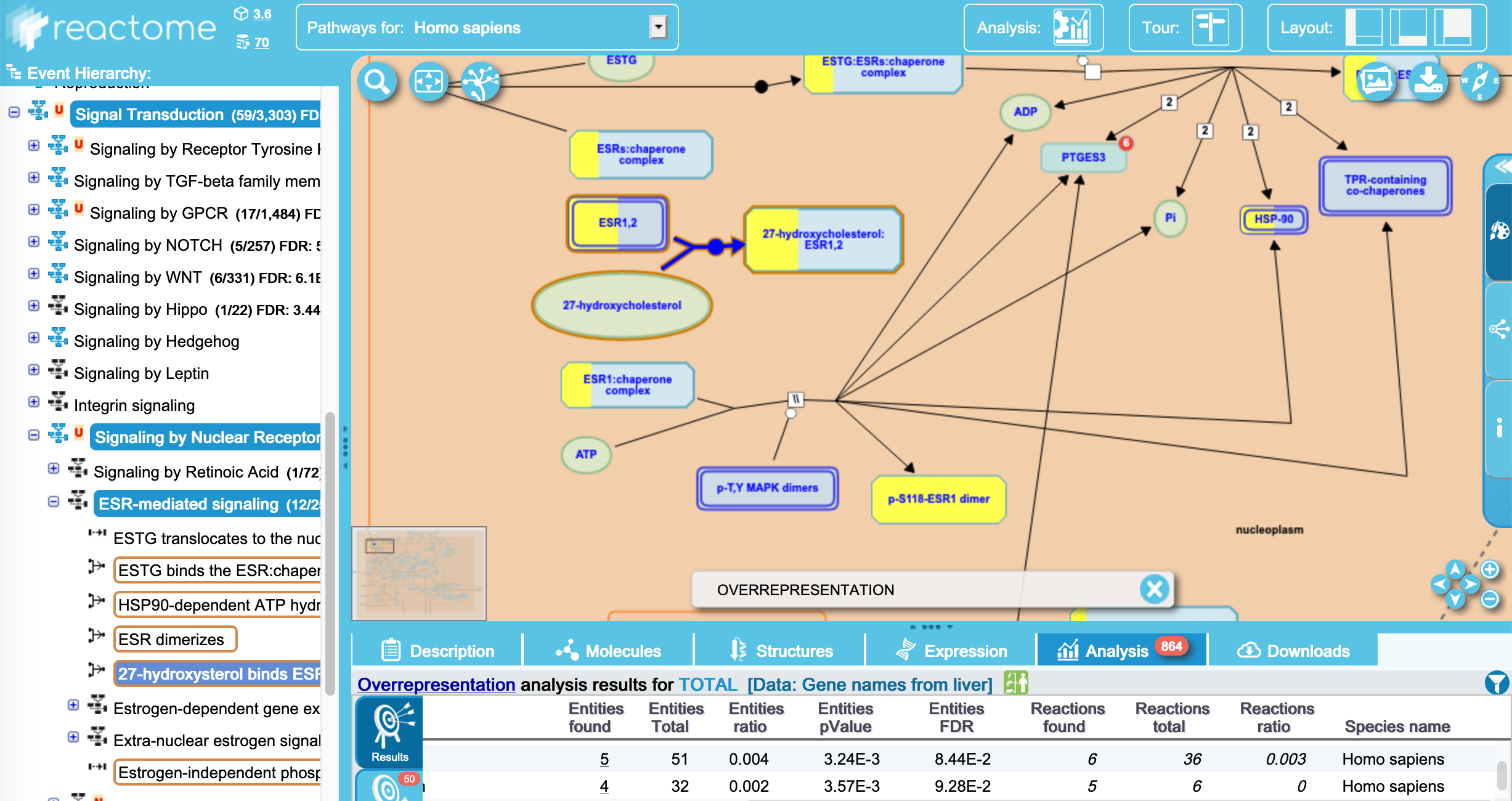
Figure 10.5: Reactome
10.6 Biocyc
Biocyc is another suite of tools for enrichment and pathway browsing, which is particularly useful for prokaryotic work. It is licensed, but Monash does have a license.
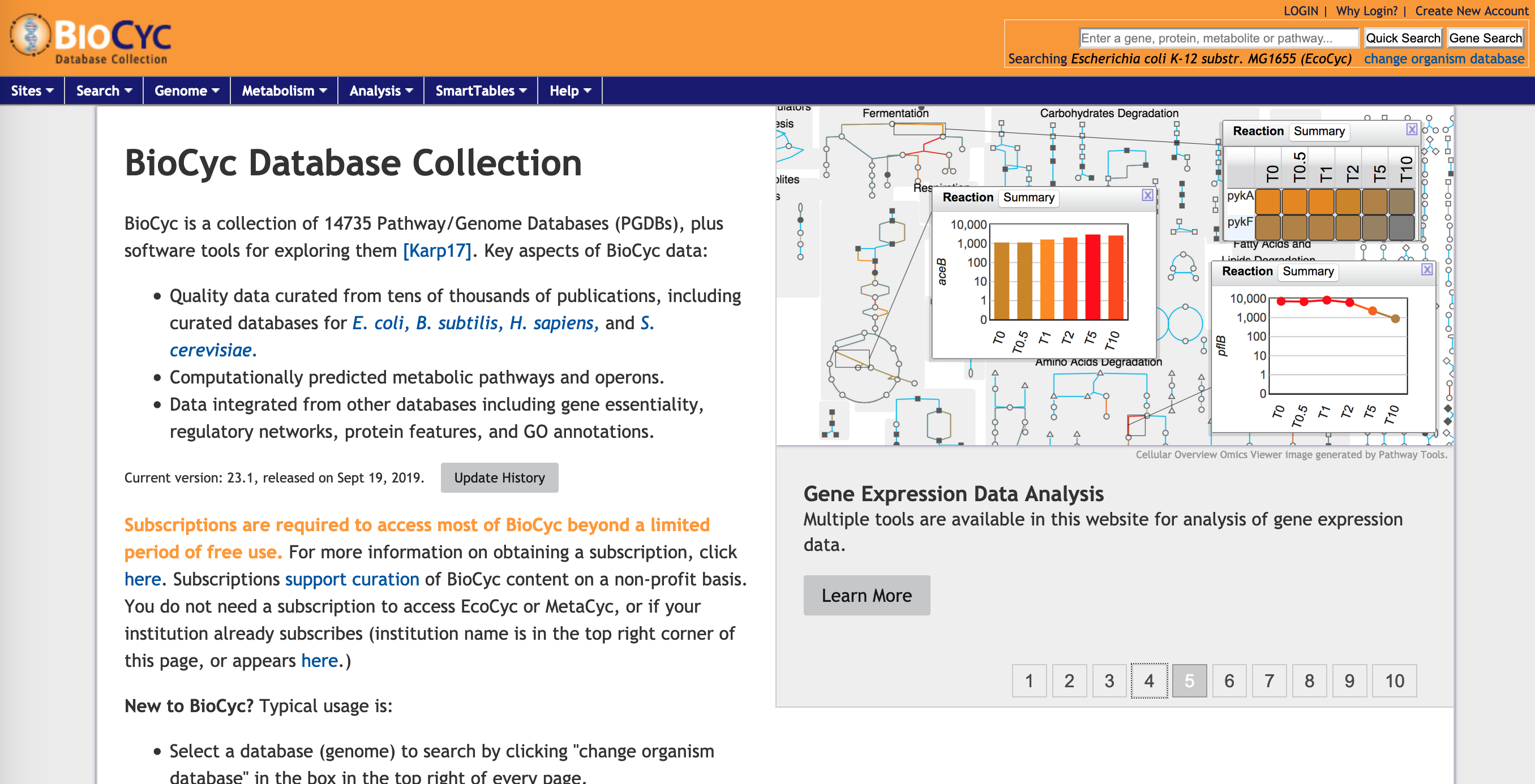
Figure 10.6: Biocyc
10.7 STRING
STRING was originally designed as a convenient tool to explore interactions within a gene list, visualised as an interaction network. While it is best suited for smaller gene lists, it now also includes functionality for performing functional enrichment analysis.
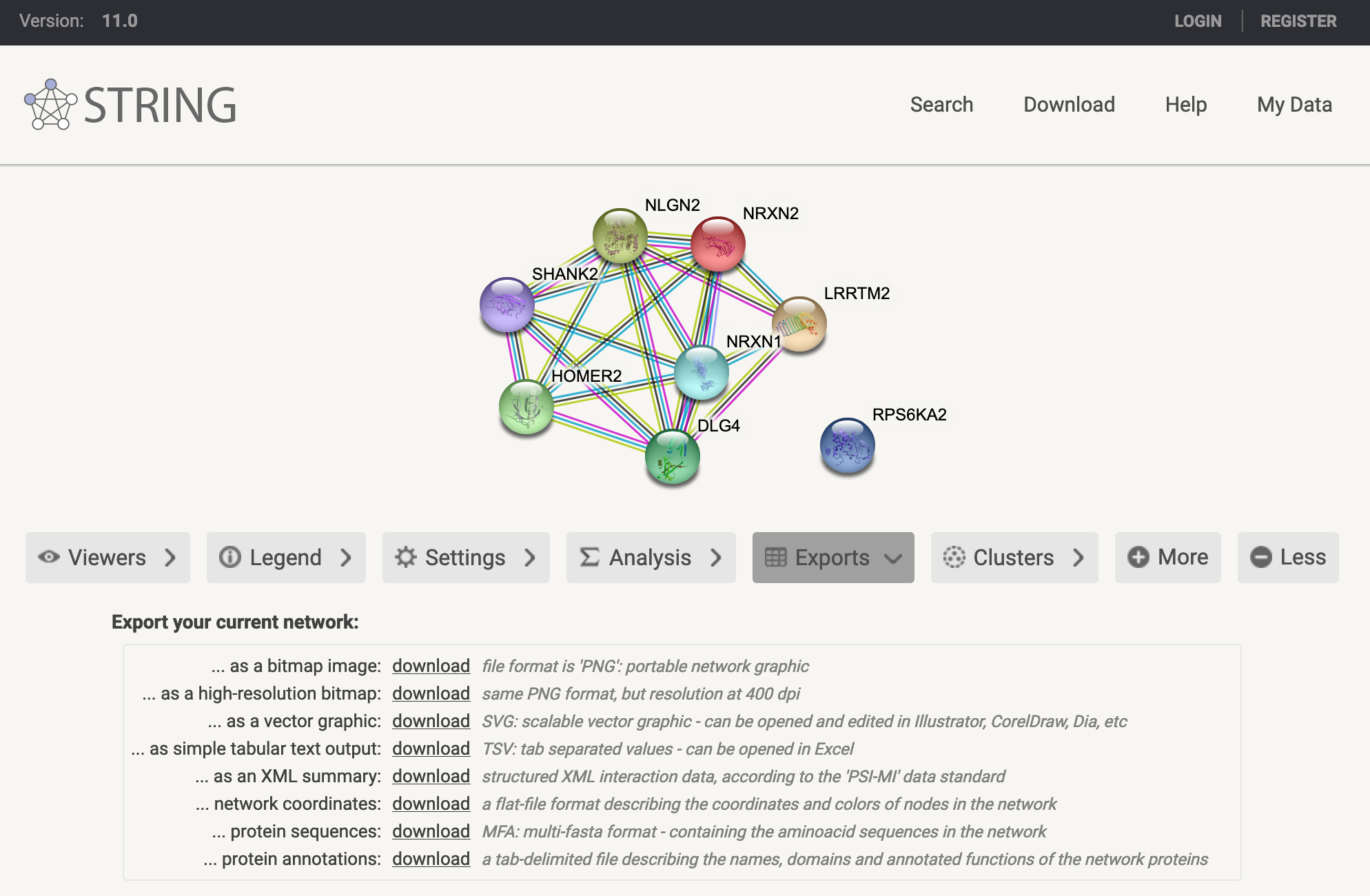
Figure 10.7: STRING
10.8 Gene Ontology
Gene Ontology (GO) terms are the most widely use set of functional annotations, used by many enrichment tools. The gene ontology resource website itself provides several tools for browsing the GO term hierarchy.
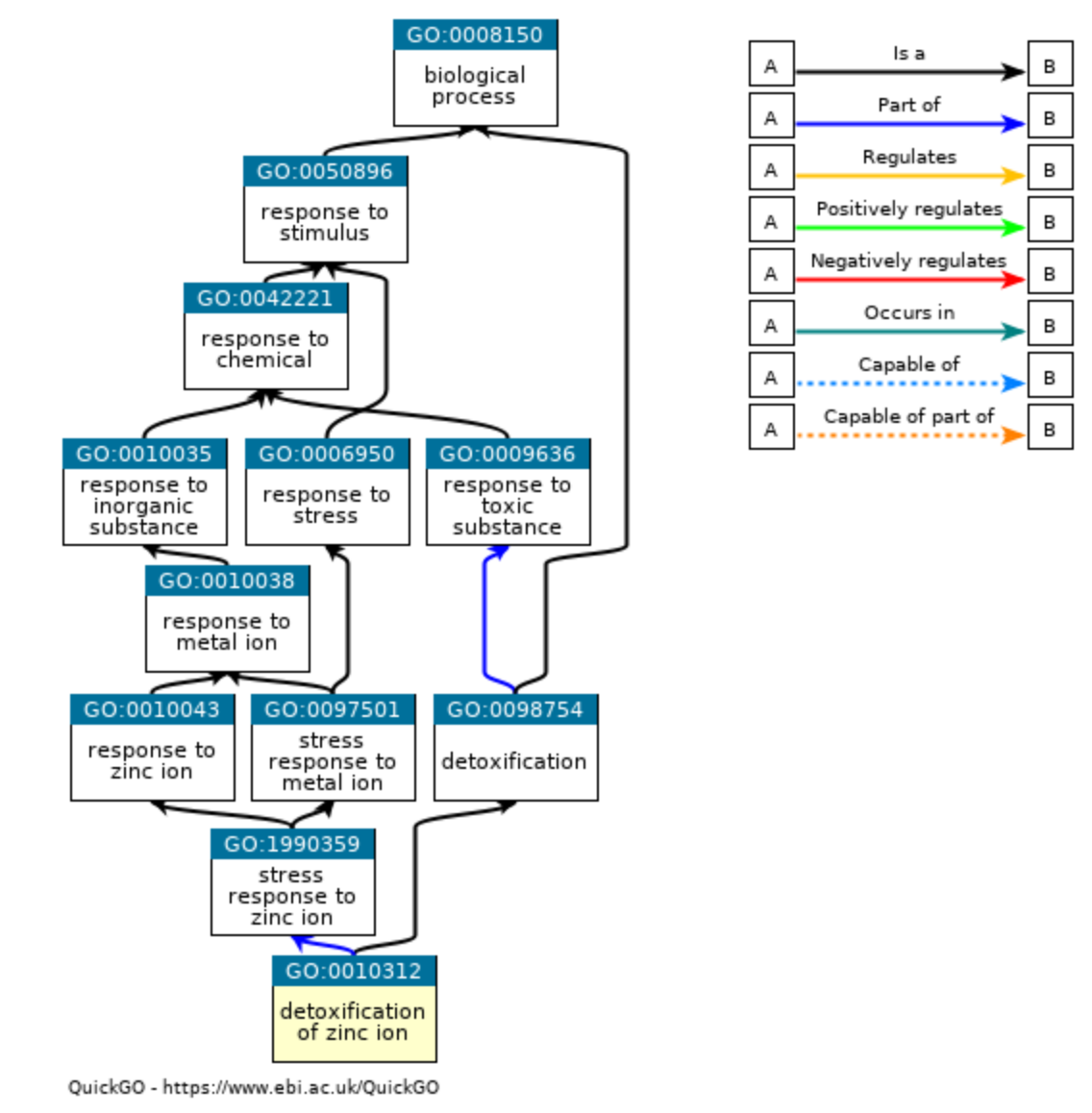
Figure 10.8: Gene Ontology
10.9 KEGG
A well known curated pathway database. It is used by many other tools but with a caveat - KEGG moved to a subscription model in 2011, and so enrichment tools need to use the last open release from 2011. However up to date KEGG pathways are browsable directly through their website.
Figure 10.9: KEGG
10.10 GSEA and MSigDB
http://software.broadinstitute.org/gsea/index.jsp
The GSEA Desktop is (one of many) gene set enrichment approaches. It uses of gene rankings across all genes rather than hypogeometric or fishers-exact tests of gene list enrichment. MSigDB (Molecular signatures database) is a suite of annotation databases suitable for GSEA analysis.
Figure 10.10: MSigDB
10.11 MetaboAnalyst
MetaboAnalyst is popular among the metabolomics community for statistical, functional and integrative analysis of metabolomics data. It has a feature called Functional enrichment analysis, which performs metabolite set enrichment analysis, metabolic pathway analysis, and pathway activity prediction from MS peaks.
Figure 10.11: MetaboAnalyst
10.12 Cytoscape
Cytoscape is a desktop-based biological network analysis / visualisation tool, rather than a functional enrichment tool (although plugins can change that). It is mentioned here because it is often useful as a next step when you need to create custom figures showing the interactions of an interesting biological pathway.

Figure 10.12: Cytoscape