13 Differential Expression
There are many different methods for calculating differential expression between groups in scRNAseq data. There are a number of review papers worth consulting on this topic.
There is the Seurat differential expression Vignette which walks through the variety implemented in Seurat.
There is also a good discussion of useing pseudobulk approaches which is worth checking out if youre planning differential expression analyses.
head(seurat_object@meta.data)
#> orig.ident nCount_RNA nFeature_RNA ind
#> AGGGCGCTATTTCC-1 SeuratProject 2053 532 1256
#> GGAGACGATTCGTT-1 SeuratProject 881 392 1256
#> CACCGTTGTCGTAG-1 SeuratProject 3130 1005 1016
#> TATCGTACACGCAT-1 SeuratProject 1042 549 1488
#> TACGAGACCTATTC-1 SeuratProject 2425 777 1244
#> GTACTACTCATACG-1 SeuratProject 3951 1064 1256
#> stim cell multiplets
#> AGGGCGCTATTTCC-1 stim CD14+ Monocytes singlet
#> GGAGACGATTCGTT-1 stim CD4 T cells singlet
#> CACCGTTGTCGTAG-1 ctrl FCGR3A+ Monocytes singlet
#> TATCGTACACGCAT-1 stim B cells singlet
#> TACGAGACCTATTC-1 stim CD4 T cells singlet
#> GTACTACTCATACG-1 ctrl FCGR3A+ Monocytes singlet
#> percent.mt RNA_snn_res.0.5 seurat_clusters
#> AGGGCGCTATTTCC-1 1.6336634 1 0
#> GGAGACGATTCGTT-1 4.8809524 6 15
#> CACCGTTGTCGTAG-1 1.0655473 4 11
#> TATCGTACACGCAT-1 3.0662710 3 4
#> TACGAGACCTATTC-1 1.0837849 0 8
#> GTACTACTCATACG-1 0.7137395 4 11
#> pca_clusters harmony_clusters
#> AGGGCGCTATTTCC-1 3 1
#> GGAGACGATTCGTT-1 0 6
#> CACCGTTGTCGTAG-1 9 4
#> TATCGTACACGCAT-1 6 3
#> TACGAGACCTATTC-1 0 0
#> GTACTACTCATACG-1 9 4
#> RNA_snn_res.0.1 RNA_snn_res.0.2
#> AGGGCGCTATTTCC-1 1 1
#> GGAGACGATTCGTT-1 0 0
#> CACCGTTGTCGTAG-1 4 4
#> TATCGTACACGCAT-1 3 3
#> TACGAGACCTATTC-1 0 0
#> GTACTACTCATACG-1 4 4
#> RNA_snn_res.0.3 RNA_snn_res.0.4
#> AGGGCGCTATTTCC-1 1 1
#> GGAGACGATTCGTT-1 0 6
#> CACCGTTGTCGTAG-1 4 4
#> TATCGTACACGCAT-1 3 3
#> TACGAGACCTATTC-1 0 0
#> GTACTACTCATACG-1 4 4
#> RNA_snn_res.0.6 RNA_snn_res.0.7
#> AGGGCGCTATTTCC-1 1 1
#> GGAGACGATTCGTT-1 7 7
#> CACCGTTGTCGTAG-1 5 6
#> TATCGTACACGCAT-1 3 4
#> TACGAGACCTATTC-1 0 3
#> GTACTACTCATACG-1 5 6
#> RNA_snn_res.0.8 RNA_snn_res.0.9
#> AGGGCGCTATTTCC-1 1 1
#> GGAGACGATTCGTT-1 9 9
#> CACCGTTGTCGTAG-1 7 7
#> TATCGTACACGCAT-1 3 3
#> TACGAGACCTATTC-1 0 0
#> GTACTACTCATACG-1 7 7
#> RNA_snn_res.1 RNA_snn_res.1.1
#> AGGGCGCTATTTCC-1 0 1
#> GGAGACGATTCGTT-1 9 8
#> CACCGTTGTCGTAG-1 8 7
#> TATCGTACACGCAT-1 3 4
#> TACGAGACCTATTC-1 6 0
#> GTACTACTCATACG-1 8 7
#> RNA_snn_res.1.2 RNA_snn_res.1.3
#> AGGGCGCTATTTCC-1 0 2
#> GGAGACGATTCGTT-1 10 11
#> CACCGTTGTCGTAG-1 8 9
#> TATCGTACACGCAT-1 3 3
#> TACGAGACCTATTC-1 2 7
#> GTACTACTCATACG-1 8 9
#> RNA_snn_res.1.4 RNA_snn_res.1.5
#> AGGGCGCTATTTCC-1 0 1
#> GGAGACGATTCGTT-1 12 12
#> CACCGTTGTCGTAG-1 10 10
#> TATCGTACACGCAT-1 1 3
#> TACGAGACCTATTC-1 2 4
#> GTACTACTCATACG-1 10 10
#> RNA_snn_res.1.6 RNA_snn_res.1.7
#> AGGGCGCTATTTCC-1 1 0
#> GGAGACGATTCGTT-1 12 13
#> CACCGTTGTCGTAG-1 10 11
#> TATCGTACACGCAT-1 2 1
#> TACGAGACCTATTC-1 9 7
#> GTACTACTCATACG-1 10 11
#> RNA_snn_res.1.8 RNA_snn_res.1.9
#> AGGGCGCTATTTCC-1 0 7
#> GGAGACGATTCGTT-1 13 14
#> CACCGTTGTCGTAG-1 11 12
#> TATCGTACACGCAT-1 1 0
#> TACGAGACCTATTC-1 8 8
#> GTACTACTCATACG-1 11 12
#> RNA_snn_res.2 Naive_CD4_T1 cell_label
#> AGGGCGCTATTTCC-1 0 -0.3540023 <NA>
#> GGAGACGATTCGTT-1 15 2.1376158 Naive_CD4_T
#> CACCGTTGTCGTAG-1 11 -1.1487836 <NA>
#> TATCGTACACGCAT-1 4 0.5557941 <NA>
#> TACGAGACCTATTC-1 8 1.4250065 Naive_CD4_T
#> GTACTACTCATACG-1 11 -0.2082793 <NA>
#> SingleR.labels
#> AGGGCGCTATTTCC-1 Monocyte
#> GGAGACGATTCGTT-1 T_cells
#> CACCGTTGTCGTAG-1 Monocyte
#> TATCGTACACGCAT-1 Neutrophils
#> TACGAGACCTATTC-1 T_cells
#> GTACTACTCATACG-1 MonocyteHow cells from each condition do we have?
table(seurat_object$stim)
#>
#> ctrl stim
#> 2378 2499How many cells per individuals per group?
table(seurat_object$ind, seurat_object$stim)
#>
#> ctrl stim
#> 101 175 226
#> 107 115 106
#> 1015 503 489
#> 1016 335 348
#> 1039 87 100
#> 1244 373 311
#> 1256 384 385
#> 1488 406 534And for each sample, how many of each cell type has been classified?
table(paste(seurat_object$ind,seurat_object$stim), seurat_object$cell)
#>
#> B cells CD14+ Monocytes CD4 T cells CD8 T cells
#> 101 ctrl 23 47 60 15
#> 101 stim 30 52 83 17
#> 1015 ctrl 79 149 142 44
#> 1015 stim 68 149 148 21
#> 1016 ctrl 21 71 82 107
#> 1016 stim 29 65 65 109
#> 1039 ctrl 7 32 36 6
#> 1039 stim 6 26 48 5
#> 107 ctrl 9 50 32 6
#> 107 stim 9 35 43 1
#> 1244 ctrl 23 85 202 8
#> 1244 stim 18 58 191 4
#> 1256 ctrl 32 80 176 26
#> 1256 stim 42 70 194 25
#> 1488 ctrl 36 59 246 13
#> 1488 stim 59 59 319 15
#>
#> Dendritic cells FCGR3A+ Monocytes
#> 101 ctrl 4 11
#> 101 stim 6 23
#> 1015 ctrl 3 49
#> 1015 stim 17 43
#> 1016 ctrl 4 21
#> 1016 stim 2 32
#> 1039 ctrl 1 3
#> 1039 stim 1 6
#> 107 ctrl 3 12
#> 107 stim 2 5
#> 1244 ctrl 8 19
#> 1244 stim 6 4
#> 1256 ctrl 6 20
#> 1256 stim 3 11
#> 1488 ctrl 8 25
#> 1488 stim 12 28
#>
#> Megakaryocytes NK cells
#> 101 ctrl 4 11
#> 101 stim 1 14
#> 1015 ctrl 5 32
#> 1015 stim 5 38
#> 1016 ctrl 3 26
#> 1016 stim 1 45
#> 1039 ctrl 1 1
#> 1039 stim 3 5
#> 107 ctrl 0 3
#> 107 stim 0 11
#> 1244 ctrl 3 25
#> 1244 stim 4 26
#> 1256 ctrl 1 43
#> 1256 stim 7 33
#> 1488 ctrl 4 15
#> 1488 stim 6 3613.1 Prefiltering
Why do we need to do this?
If expression is below a certain level, it will be almost impossible to see any differential expression.
When doing differential expression, you generally ignore genes with low expression. In single cell datasets, there are many genes like this. Filtering here to make our dataset smaller so it runs quicker, and there is less aggressive correction for multiple hypotheses.
How many genes before filtering?
seurat_object
#> An object of class Seurat
#> 35635 features across 4877 samples within 1 assay
#> Active assay: RNA (35635 features, 2000 variable features)
#> 3 layers present: counts, data, scale.data
#> 4 dimensional reductions calculated: pca, umap, harmony, umap_harmonyHow many copies of each gene are there?
total_per_gene <- rowSums(GetAssayData(seurat_object, assay='RNA', slot='counts'))
#> Warning: The `slot` argument of `GetAssayData()` is deprecated as of SeuratObject 5.0.0.
#> ℹ Please use the `layer` argument instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.
hist(log10(total_per_gene))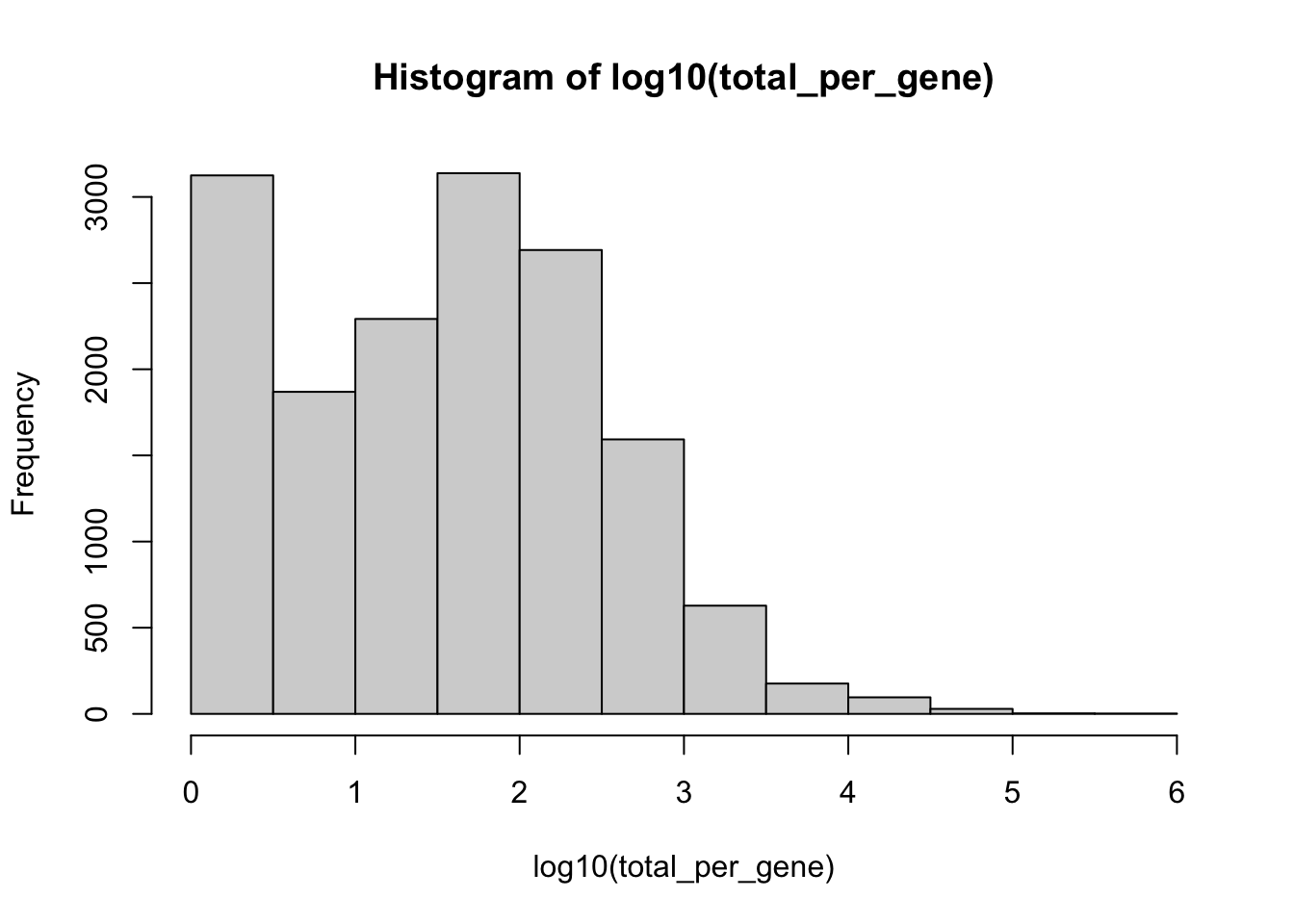
Lets keep only those genes with at least 50 copies across the entire experiment.
seurat_object<- seurat_object[total_per_gene >= 50, ] How many genes after filtering?
seurat_object
#> An object of class Seurat
#> 7200 features across 4877 samples within 1 assay
#> Active assay: RNA (7200 features, 1236 variable features)
#> 3 layers present: counts, data, scale.data
#> 4 dimensional reductions calculated: pca, umap, harmony, umap_harmonyWe might like to see the effect of IFN-beta stimulation on each cell type individually. For the purposes of this workshop, just going to test one cell type; CD14+ Monocytes
An easy way is to subset the object.
# Set idents to 'cell' column.
Idents(seurat_object) <- seurat_object$cell
DimPlot(seurat_object, reduction = "umap_harmony")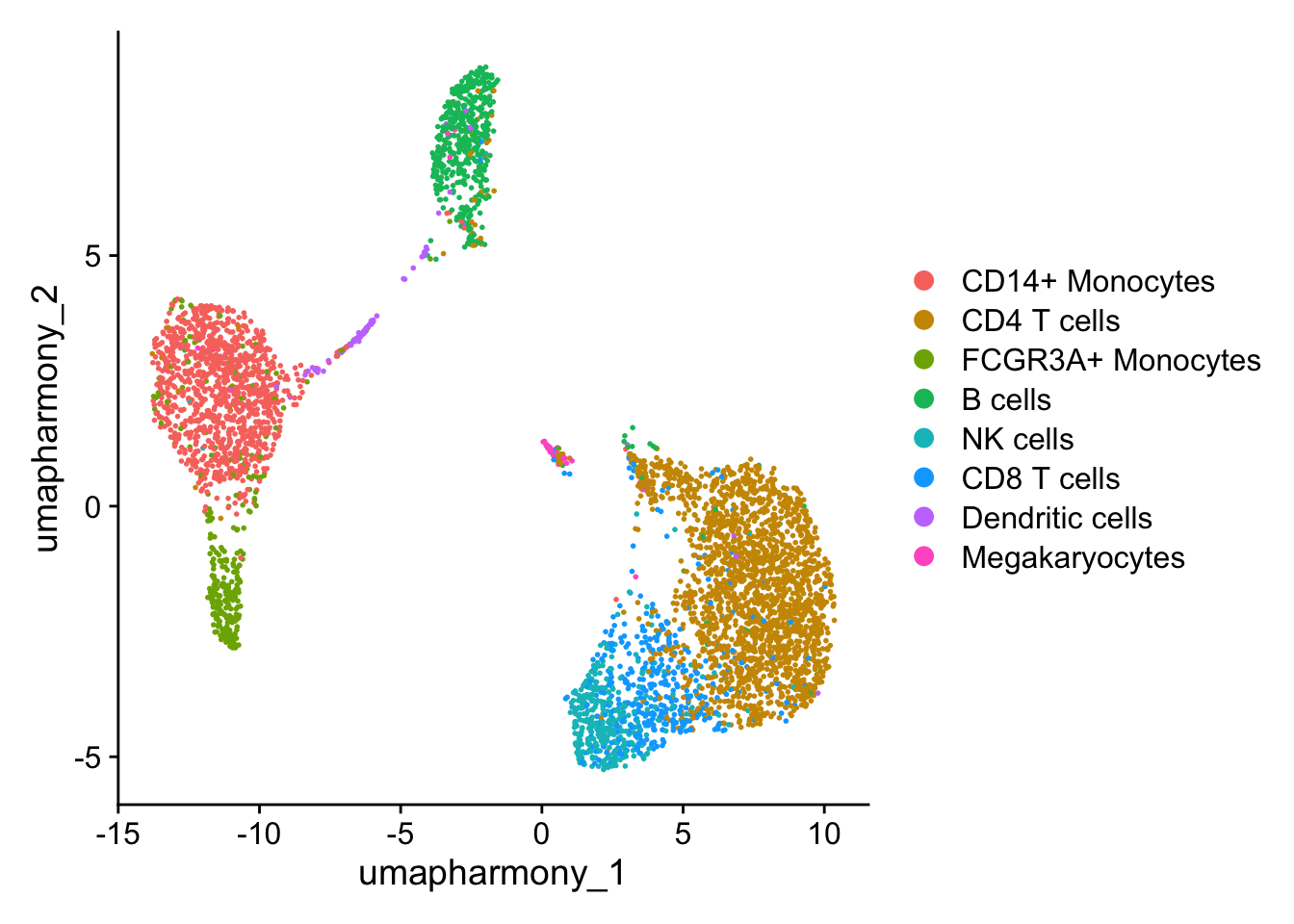
seurat_object_celltype <- seurat_object[, seurat_object$cell == "CD14+ Monocytes" ]
DimPlot(seurat_object_celltype, reduction = "umap_harmony")
13.2 Default Wilcox test
To run this test, we change the Idents to the factor(column) we want to test. In this case, that’s ‘stim’.
# Change Ident to Condition
Idents(seurat_object_celltype) <- seurat_object_celltype$stim
# default, wilcox test
de_result_wilcox <- FindMarkers(seurat_object_celltype,
ident.1 = 'stim',
ident.2 = 'ctrl',
logfc.threshold = 0, # Give me ALL results
min.pct = 0
)
# Add average expression for plotting
de_result_wilcox$AveExpr<- rowMeans(seurat_object_celltype[rownames(de_result_wilcox),])Look at the top differentially expressed genes.
head(de_result_wilcox)
#> p_val avg_log2FC pct.1 pct.2 p_val_adj
#> RSAD2 8.471603e-196 6.797508 0.979 0.044 6.099554e-192
#> TNFSF10 3.399691e-195 6.559853 0.982 0.056 2.447777e-191
#> CXCL10 5.809758e-195 8.034949 0.975 0.038 4.183025e-191
#> IFIT3 7.214495e-195 6.234703 0.982 0.051 5.194437e-191
#> IFIT1 1.224398e-188 6.764699 0.953 0.026 8.815662e-185
#> ISG15 1.228738e-186 6.377243 1.000 0.323 8.846914e-183
#> AveExpr
#> RSAD2 0.04196286
#> TNFSF10 0.51655832
#> CXCL10 3.25184139
#> IFIT3 0.02032398
#> IFIT1 0.01254584
#> ISG15 0.14165156
p1 <- ggplot(de_result_wilcox, aes(x=AveExpr, y=avg_log2FC, col=p_val_adj < 0.05)) +
geom_point() +
scale_colour_manual(values=c('TRUE'="red",'FALSE'="black")) +
theme_bw() +
ggtitle("Wilcox Test")
p2 <- ggplot(de_result_wilcox, aes(x=avg_log2FC, y=-log10(p_val), col=p_val_adj < 0.05)) +
geom_point() +
scale_colour_manual(values=c('TRUE'="red",'FALSE'="black")) +
theme_bw() +
ggtitle("Wilcox Test (Volcano)")
p1 + p2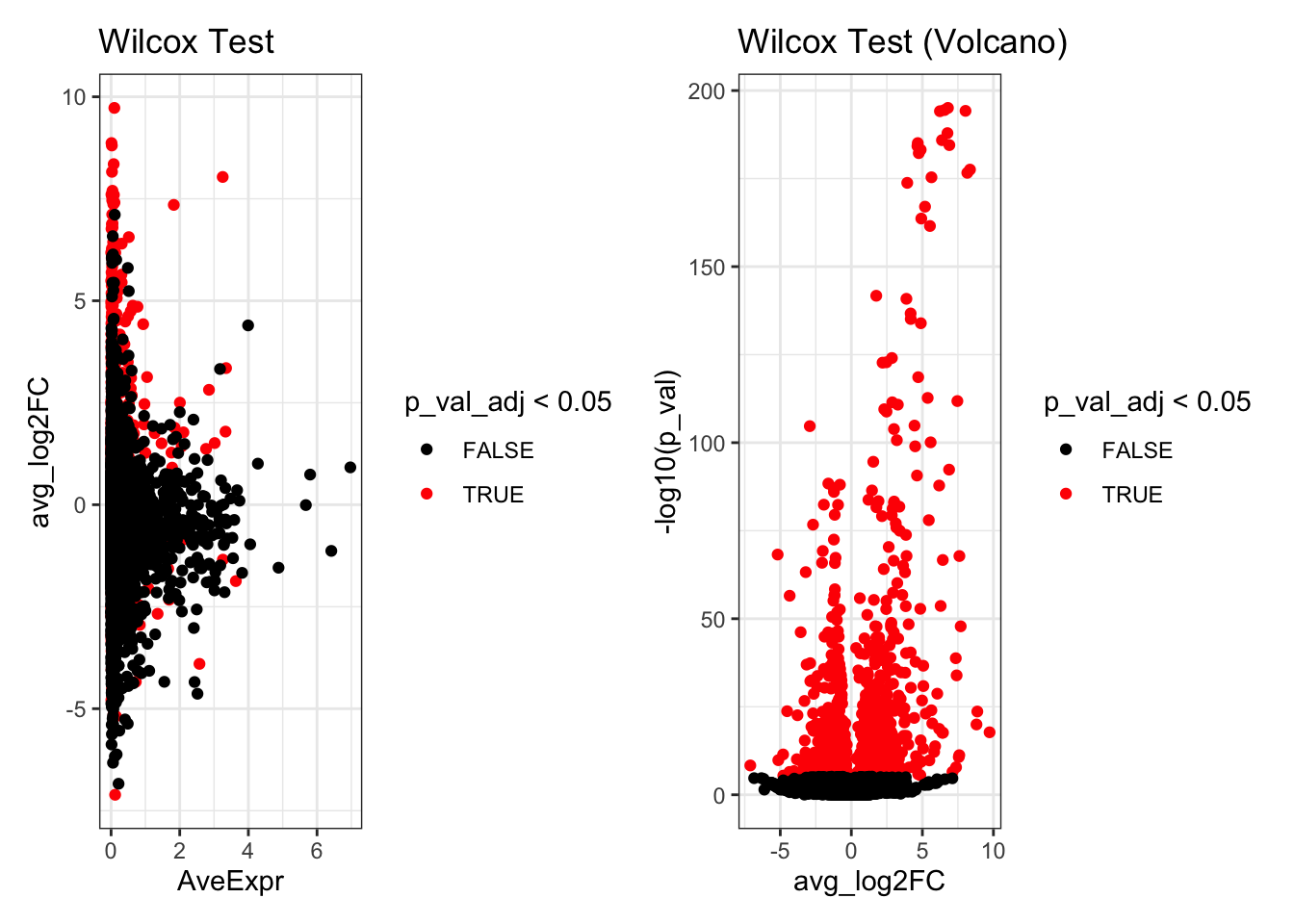
13.3 Seurat Negative binomial
Negative binonial test is run almost the same way - just need to specify it under ‘test.use’
# Change Ident to Condition
Idents(seurat_object_celltype) <- seurat_object_celltype$stim
# default, wilcox test
de_result_negbinom <- FindMarkers(seurat_object_celltype,
test.use="negbinom", # Choose a different test.
ident.1 = 'stim',
ident.2 = 'ctrl',
logfc.threshold = 0, # Give me ALL results
min.pct = 0
)
# Add average expression for plotting
de_result_negbinom$AveExpr<- rowMeans(seurat_object_celltype[rownames(de_result_negbinom),])Look at the top differentially expressed genes.
head(de_result_negbinom)
#> p_val avg_log2FC pct.1 pct.2 p_val_adj AveExpr
#> CXCL10 0 8.034949 0.975 0.038 0 0.04196286
#> CCL8 0 8.160755 0.912 0.023 0 0.51655832
#> LY6E 0 4.880093 0.979 0.134 0 3.25184139
#> APOBEC3A 0 4.745713 0.996 0.262 0 0.02032398
#> IFITM3 0 4.675898 0.998 0.271 0 0.01254584
#> IFI6 0 3.935122 0.982 0.257 0 0.14165156
p1 <- ggplot(de_result_negbinom, aes(x=AveExpr, y=avg_log2FC, col=p_val_adj < 0.05)) +
geom_point() +
scale_colour_manual(values=c('TRUE'="red",'FALSE'="black")) +
theme_bw() +
ggtitle("Negative Bionomial Test")
p2 <- ggplot(de_result_negbinom, aes(x=avg_log2FC, y=-log10(p_val), col=p_val_adj < 0.05)) +
geom_point() +
scale_colour_manual(values=c('TRUE'="red",'FALSE'="black")) +
theme_bw() +
ggtitle("Negative Bionomial Test (Volcano)")
p1 + p2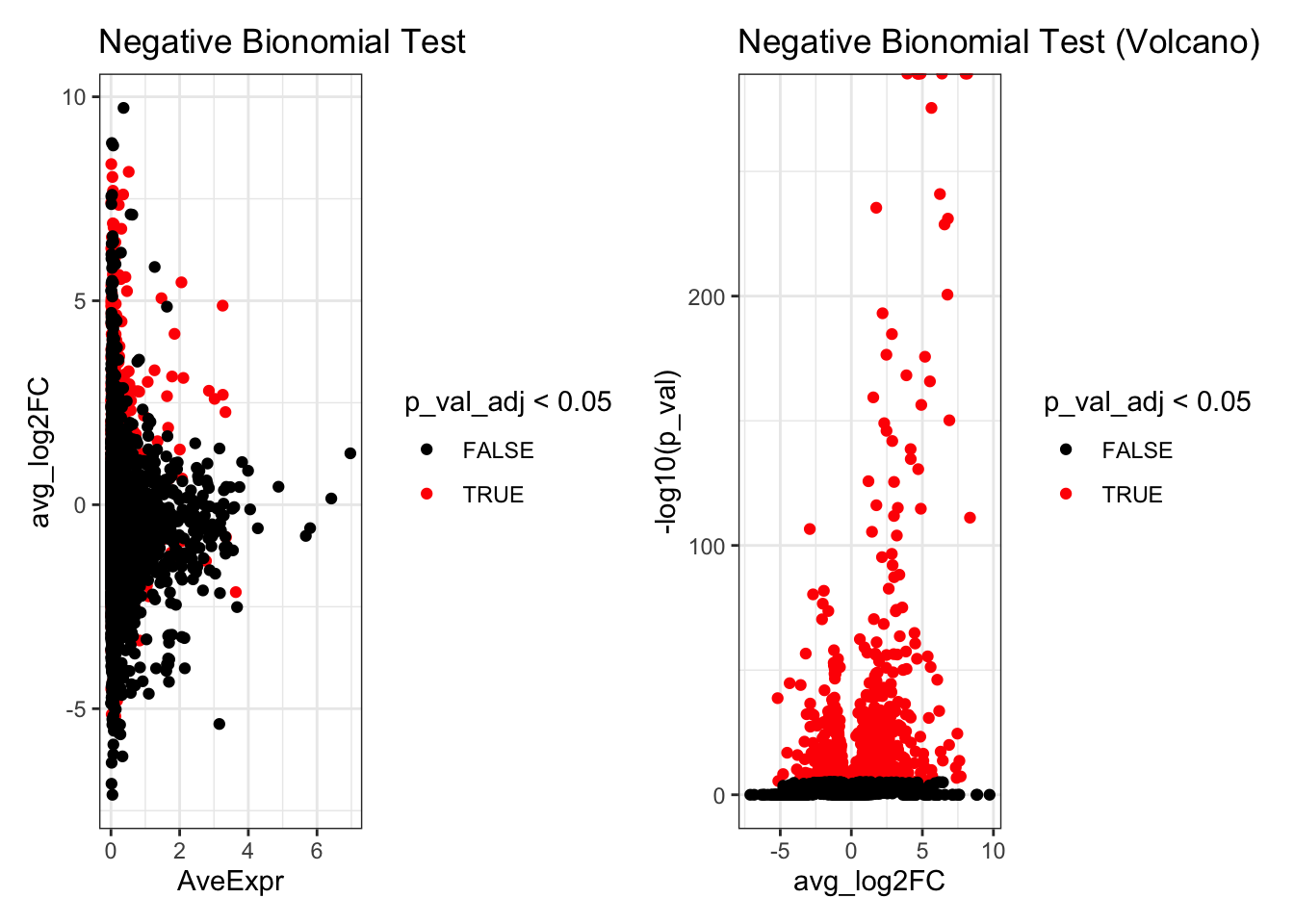
13.4 Pseudobulk
Pseudobulk analysis is an option where you have biological replicates. It is essentially pooling the individual cell counts and treating your expreiment like a bulk RNAseq.
First, you need to build a pseudobulk matrix - the AggregateExpression() function can do this, once you set the ‘Idents’ of your seurat object to your grouping factor (here, thats a combination of individual+treatment called ‘sample’, instead of the ‘stim’ treatment column).
# Tools for bulk differential expression
library(limma)
#>
#> Attaching package: 'limma'
#> The following object is masked from 'package:BiocGenerics':
#>
#> plotMA
library(edgeR)
#>
#> Attaching package: 'edgeR'
#> The following object is masked from 'package:SingleCellExperiment':
#>
#> cpm
# Change idents to ind for grouping.
seurat_object_celltype$sample <- factor(paste(seurat_object_celltype$stim, seurat_object_celltype$ind, sep="_"))
Idents(seurat_object_celltype) <- seurat_object_celltype$sample
# THen pool together counts in those groups
# AggregateExperssion returns a list of matricies - one for each assay requested (even just requesting one)
pseudobulk_matrix_list <- AggregateExpression( seurat_object_celltype, slot = 'counts', assays='RNA' )
#> Names of identity class contain underscores ('_'), replacing with dashes ('-')
#> This message is displayed once every 8 hours.
pseudobulk_matrix <- pseudobulk_matrix_list[['RNA']]
pseudobulk_matrix[1:5,1:4]
#> 5 x 4 sparse Matrix of class "dgCMatrix"
#> ctrl-101 ctrl-1015 ctrl-1016 ctrl-1039
#> NOC2L 2 7 . .
#> HES4 . 3 2 1
#> ISG15 31 185 234 41
#> TNFRSF18 . 3 4 2
#> TNFRSF4 . 2 . .Now it looks like a bulk RNAseq experiment, so treat it like one.
We can use the popular limma package for differential expression. Here is one tutorial, and the hefty reference manual is hosted by bioconductor.
In brief, this code below constructs a linear model for this experiment that accounts for the variation in individuals and treatment. It then tests for differential expression between ‘stim’ and ‘ctrl’ groups.
dge <- DGEList(pseudobulk_matrix)
dge <- calcNormFactors(dge)
# Remove _ or - and everything after it - yeilds stim group
stim <- gsub("[-_].*","",colnames(pseudobulk_matrix))
# Removing everything before the _ or - for the individual, then converting those numerical ind explictiy to text. Else limma will treat them as numbers!
ind <- as.character(gsub(".*[-_]","",colnames(pseudobulk_matrix)))
design <- model.matrix( ~0 + stim + ind)
vm <- voom(dge, design = design, plot = FALSE)
fit <- lmFit(vm, design = design)
contrasts <- makeContrasts(stimstim - stimctrl, levels=coef(fit))
fit <- contrasts.fit(fit, contrasts)
fit <- eBayes(fit)
de_result_pseudobulk <- topTable(fit, n = Inf, adjust.method = "BH")
de_result_pseudobulk <- arrange(de_result_pseudobulk , adj.P.Val)Look at the significantly differentially expressed genes:
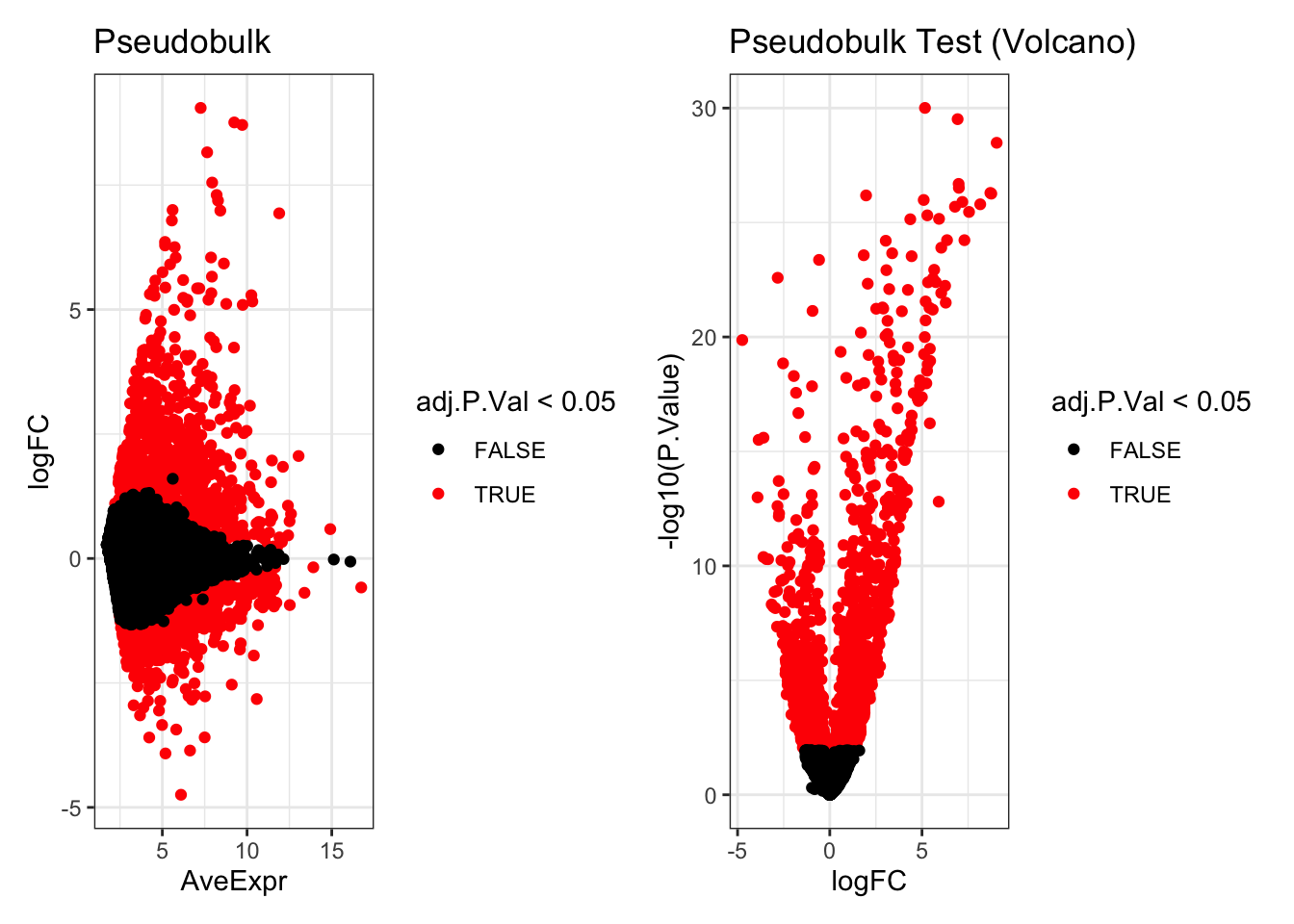
head(de_result_pseudobulk)
#> logFC AveExpr t P.Value
#> ISG20 5.161336 10.313033 35.72209 9.838370e-31
#> ISG15 6.932960 11.897783 34.63778 3.049704e-30
#> CXCL11 9.050139 7.263356 32.46019 3.282727e-29
#> IFIT3 6.987552 8.423467 28.97126 2.052443e-27
#> HERC5 6.999781 5.604886 28.65574 3.050588e-27
#> CXCL10 8.708460 9.715253 28.25406 5.081902e-27
#> adj.P.Val B
#> ISG20 7.083626e-27 60.13277
#> ISG15 1.097894e-26 59.02083
#> CXCL11 7.878545e-26 55.45779
#> IFIT3 3.694397e-24 52.01990
#> HERC5 4.392847e-24 51.18902
#> CXCL10 5.629458e-24 51.32019
p1 <- ggplot(de_result_pseudobulk, aes(x=AveExpr, y=logFC, col=adj.P.Val < 0.05)) +
geom_point() +
scale_colour_manual(values=c('TRUE'="red",'FALSE'="black")) +
theme_bw() +
ggtitle("Pseudobulk")
p2 <- ggplot(de_result_pseudobulk, aes(x=logFC, y=-log10(P.Value), col=adj.P.Val < 0.05)) +
geom_point() +
scale_colour_manual(values=c('TRUE'="red",'FALSE'="black")) +
theme_bw() +
ggtitle("Pseudobulk Test (Volcano)")
p1 + p2